ADAM User Guide¶
Introduction¶
ADAM is a library and command line tool that enables the use of Apache Spark to parallelize genomic data analysis across cluster/cloud computing environments. ADAM uses a set of schemas to describe genomic sequences, reads, variants/genotypes, and features, and can be used with data in legacy genomic file formats such as SAM/BAM/CRAM, BED/GFF3/GTF, and VCF, as well as data stored in the columnar Apache Parquet format. On a single node, ADAM provides competitive performance to optimized multi-threaded tools, while enabling scale out to clusters with more than a thousand cores. ADAM’s APIs can be used from Scala, Java, Python, R, and SQL.
The ADAM/Big Data Genomics Ecosystem¶
ADAM builds upon the open source Apache Spark, Apache Avro, and Apache Parquet projects. Additionally, ADAM can be deployed for both interactive and production workflows using a variety of platforms. A diagram of the ecosystem of tools and libraries that ADAM builds on and the tools that build upon the ADAM APIs can be found below.
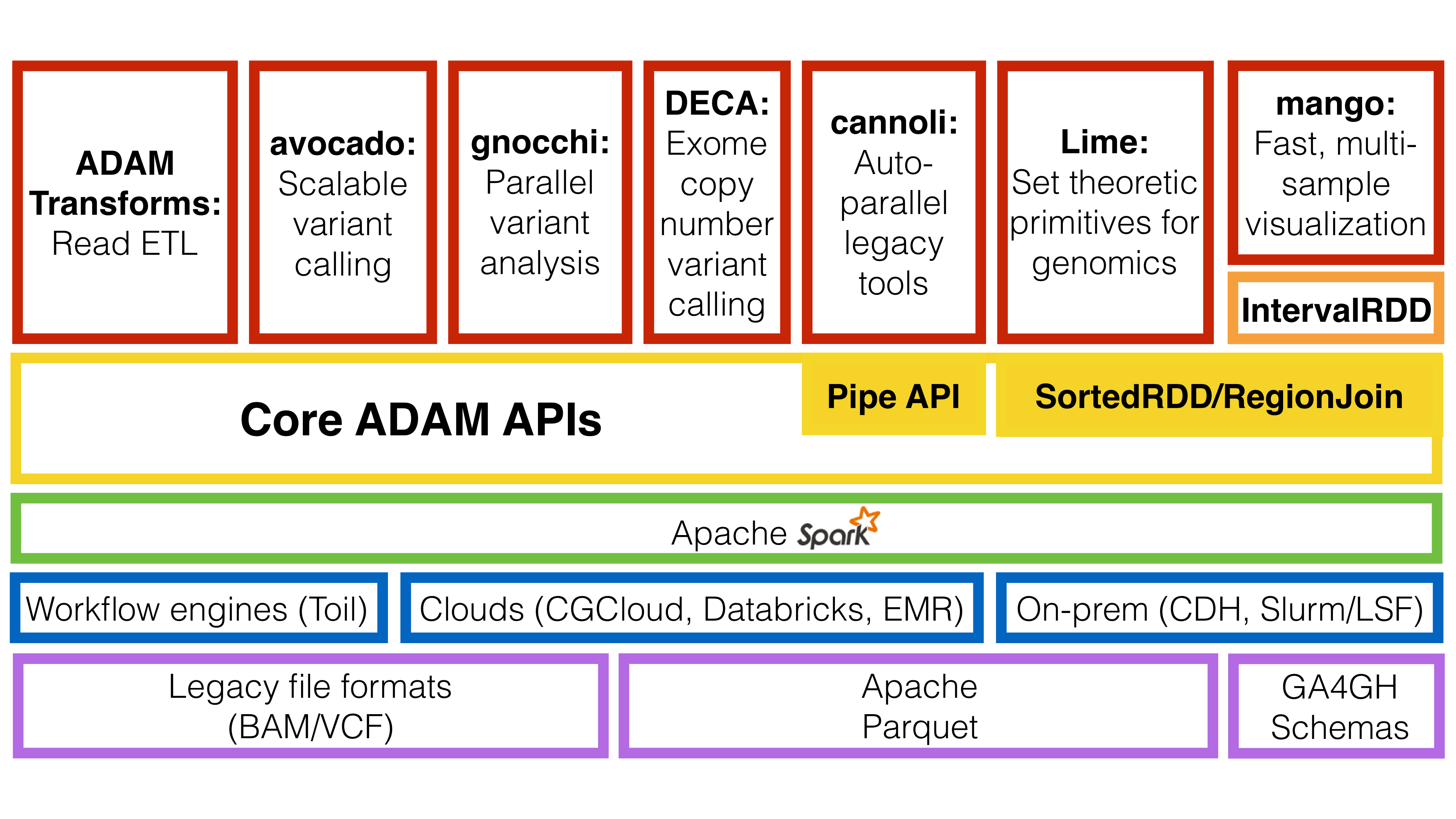
The ADAM ecosystem.
As the diagram shows, beyond the ADAM CLI, there are a number of tools built using ADAM’s core APIs:
- Avocado is a variant caller built on top of ADAM for germline and somatic calling
- Cannoli uses ADAM’s pipe API to parallelize common single-node genomics tools (e.g., BWA, bowtie2, FreeBayes)
- DECA is a reimplementation of the XHMM copy number variant caller on top of ADAM/Apache Spark
- Gnocchi provides primitives for running GWAS/eQTL tests on large genotype/phenotype datasets using ADAM
- Lime provides a parallel implementation of genomic set theoretic primitives using the region join API
- Mango is a library for visualizing large scale genomics data with interactive latencies and serving data using the GA4GH schemas
Architecture
Installation
Deploying ADAM
ADAM's APIs
Building Downstream Applications
Algorithms in ADAM
References¶
Armbrust, Michael, Reynold S. Xin, Cheng Lian, Yin Huai, Davies Liu, Joseph K. Bradley, Xiangrui Meng, et al. 2015. “Spark SQL: Relational Data Processing in Spark.” In Proceedings of the International Conference on Management of Data (SIGMOD ‘15).
DePristo, Mark A, Eric Banks, Ryan Poplin, Kiran V Garimella, Jared R Maguire, Christopher Hartl, Anthony A Philippakis, et al. 2011. “A Framework for Variation Discovery and Genotyping Using Next-Generation DNA Sequencing Data.” Nature Genetics 43 (5). Nature Publishing Group: 491–98.
Langmead, Ben, Michael C Schatz, Jimmy Lin, Mihai Pop, and Steven L Salzberg. 2009. “Searching for SNPs with Cloud Computing.” Genome Biology 10 (11). BioMed Central: R134.
Li, Heng, and Richard Durbin. 2010. “Fast and Accurate Long-Read Alignment with Burrows-Wheeler Transform.” Bioinformatics 26 (5). Oxford Univ Press: 589–95.
Massie, Matt, Frank Nothaft, Christopher Hartl, Christos Kozanitis, André Schumacher, Anthony D Joseph, and David A Patterson. 2013. “ADAM: Genomics Formats and Processing Patterns for Cloud Scale Computing.” UCB/EECS-2013-207, EECS Department, University of California, Berkeley.
McKenna, Aaron, Matthew Hanna, Eric Banks, Andrey Sivachenko, Kristian Cibulskis, Andrew Kernytsky, Kiran Garimella, et al. 2010. “The Genome Analysis Toolkit: A MapReduce Framework for Analyzing Next-Generation DNA Sequencing Data.” Genome Research 20 (9). Cold Spring Harbor Lab: 1297–1303.
Melnik, Sergey, Andrey Gubarev, Jing Jing Long, Geoffrey Romer, Shiva Shivakumar, Matt Tolton, and Theo Vassilakis. 2010. “Dremel: Interactive Analysis of Web-Scale Datasets.” Proceedings of the VLDB Endowment 3 (1-2). VLDB Endowment: 330–39.
Nakamura, Kensuke, Taku Oshima, Takuya Morimoto, Shun Ikeda, Hirofumi Yoshikawa, Yuh Shiwa, Shu Ishikawa, et al. 2011. “Sequence-Specific Error Profile of Illumina Sequencers.” Nucleic Acids Research. Oxford Univ Press, gkr344.
Nothaft, Frank A, Matt Massie, Timothy Danford, Zhao Zhang, Uri Laserson, Carl Yeksigian, Jey Kottalam, et al. 2015. “Rethinking Data-Intensive Science Using Scalable Analytics Systems.” In Proceedings of the 2015 ACM SIGMOD International Conference on Management of Data (SIGMOD ’15). ACM.
Sandberg, Russel, David Goldberg, Steve Kleiman, Dan Walsh, and Bob Lyon. 1985. “Design and Implementation of the Sun Network Filesystem.” In Proceedings of the USENIX Conference, 119–30.
Schadt, Eric E, Michael D Linderman, Jon Sorenson, Lawrence Lee, and Garry P Nolan. 2010. “Computational Solutions to Large-Scale Data Management and Analysis.” Nature Reviews Genetics 11 (9). Nature Publishing Group: 647–57.
Schatz, Michael C. 2009. “CloudBurst: Highly Sensitive Read Mapping with MapReduce.” Bioinformatics 25 (11). Oxford Univ Press: 1363–69.
Sherry, Stephen T, M-H Ward, M Kholodov, J Baker, Lon Phan, Elizabeth M Smigielski, and Karl Sirotkin. 2001. “dbSNP: The NCBI Database of Genetic Variation.” Nucleic Acids Research 29 (1). Oxford Univ Press: 308–11.
Smith, Temple F, and Michael S Waterman. 1981. “Identification of Common Molecular Subsequences.” Journal of Molecular Biology 147 (1). Elsevier: 195–97.
The Broad Institute of Harvard and MIT. 2014. “Picard.” http://broadinstitute.github.io/picard/.
Vavilapalli, Vinod Kumar, Arun C Murthy, Chris Douglas, Sharad Agarwal, Mahadev Konar, Robert Evans, Thomas Graves, et al. 2013. “Apache Hadoop YARN: Yet Another Resource Negotiator.” In Proceedings of the Symposium on Cloud Computing (SoCC ‘13), 5. ACM.
Vivian, John, Arjun Rao, Frank Austin Nothaft, Christopher Ketchum, Joel Armstrong, Adam Novak, Jacob Pfeil, et al. 2016. “Rapid and Efficient Analysis of 20,000 RNA-Seq Samples with Toil.” BioRxiv. Cold Spring Harbor Labs Journals.
Zaharia, Matei, Mosharaf Chowdhury, Tathagata Das, Ankur Dave, Justin Ma, Murphy McCauley, Michael Franklin, Scott Shenker, and Ion Stoica. 2012. “Resilient Distributed Datasets: A Fault-Tolerant Abstraction for in-Memory Cluster Computing.” In Proceedings of the Conference on Networked Systems Design and Implementation (NSDI ’12), 2. USENIX Association.
Zimmermann, Hubert. 1980. “OSI Reference Model–The ISO Model of Architecture for Open Systems Interconnection.” IEEE Transactions on Communications 28 (4). IEEE: 425–32.

