Storage Size¶
ADAM uses Apache Parquet as a way to store genomic data. This is in addition to our support for conventional genomic file formats. Parquet is an efficient columnar storage system that is widely used in the analytics ecosystem, and integrates with a variety of data management tools and query engines. Parquet provides improved storage capacity relative to several conventional genomics data storage formats. Here, we look at the storage cost of aligned reads, features, and variants.
Aligned Reads¶
In this benchmark, we have stored a copy of NA12878 aligned to the GRCh37 reference genome using BWA. We store this genome in BAM, CRAM, and ADAM, using the default compression settings for each. BAM and CRAM files were generated using htslib. This read file was sequenced at approximately 60x coverage across the whole genome.
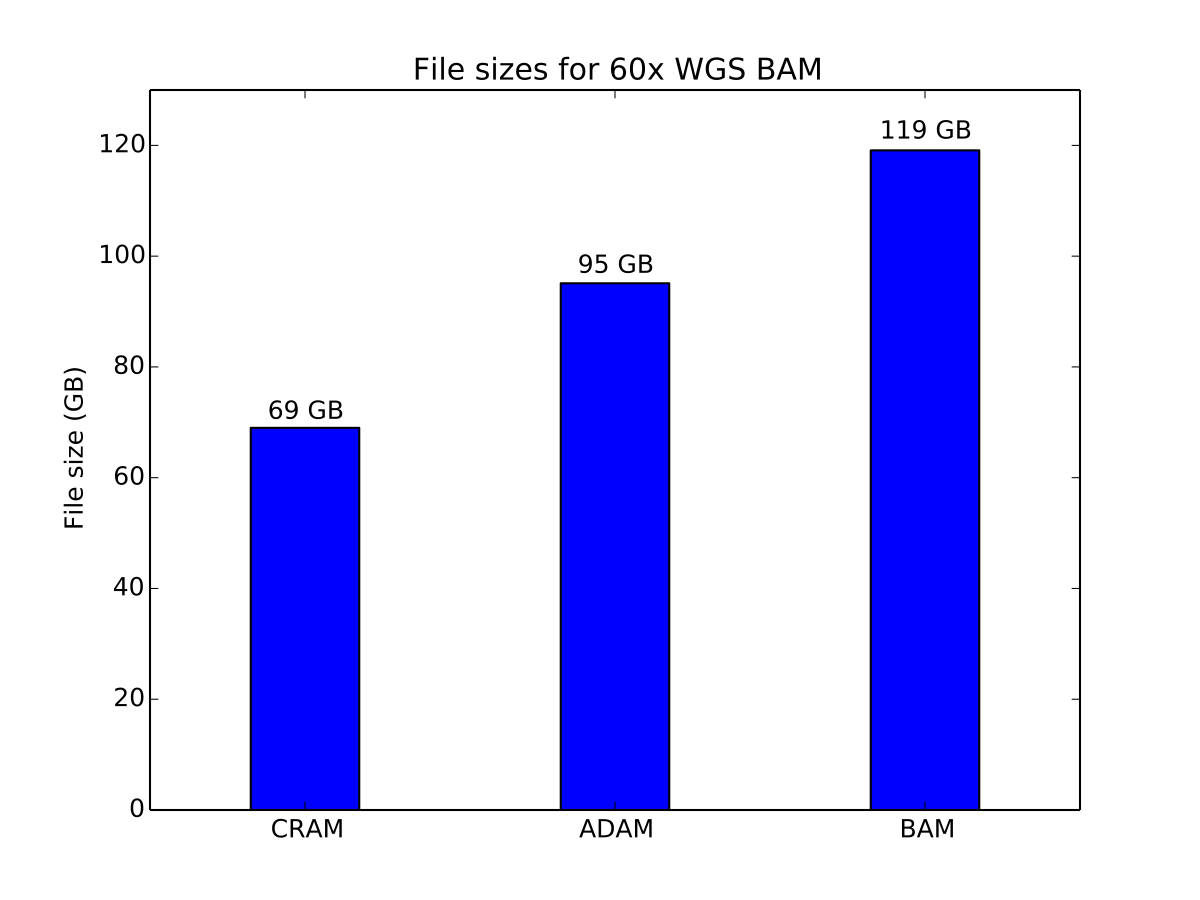
Storage cost of a 60x coverage WGS aligned dataset
ADAM provides a 20% improvement in storage size over BAM, while CRAM achieves a 43% improvement in storage cost. CRAM achieves a higher compression ratio by using reference-based compression techniques to minimize the amount of data stored on disk.
Features¶
Here, we benchmark both the GFF3 and BED formats. For GFF3, we use the ENSEMBL GRCh38 genome annotation file. For BED, we use genome-wide coverage counts generated from the NA12878 dataset used in the aligned read benchmarks.
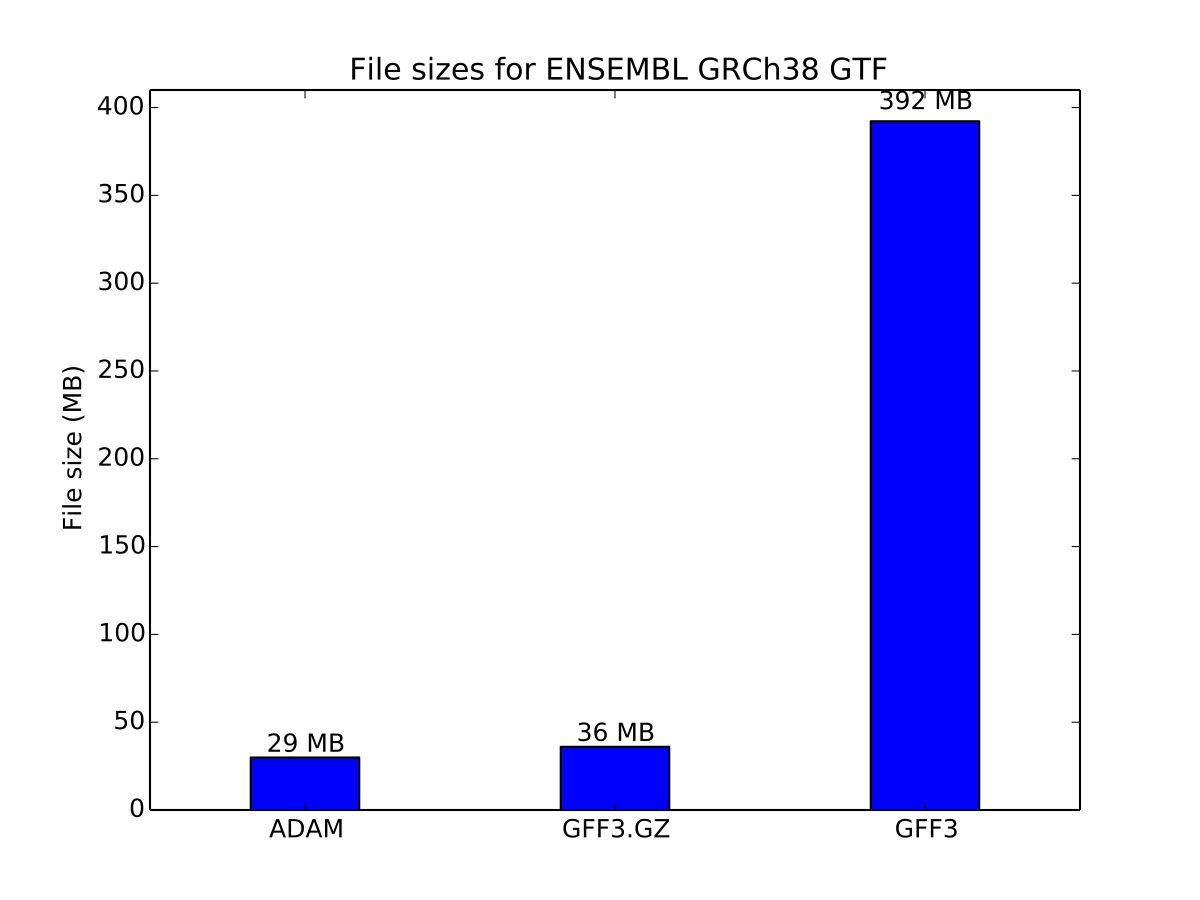
Storage cost of genome annotations
For the genome annotation file, ADAM provides a 20% improvement in storage size relative to the compressed GFF3 file.
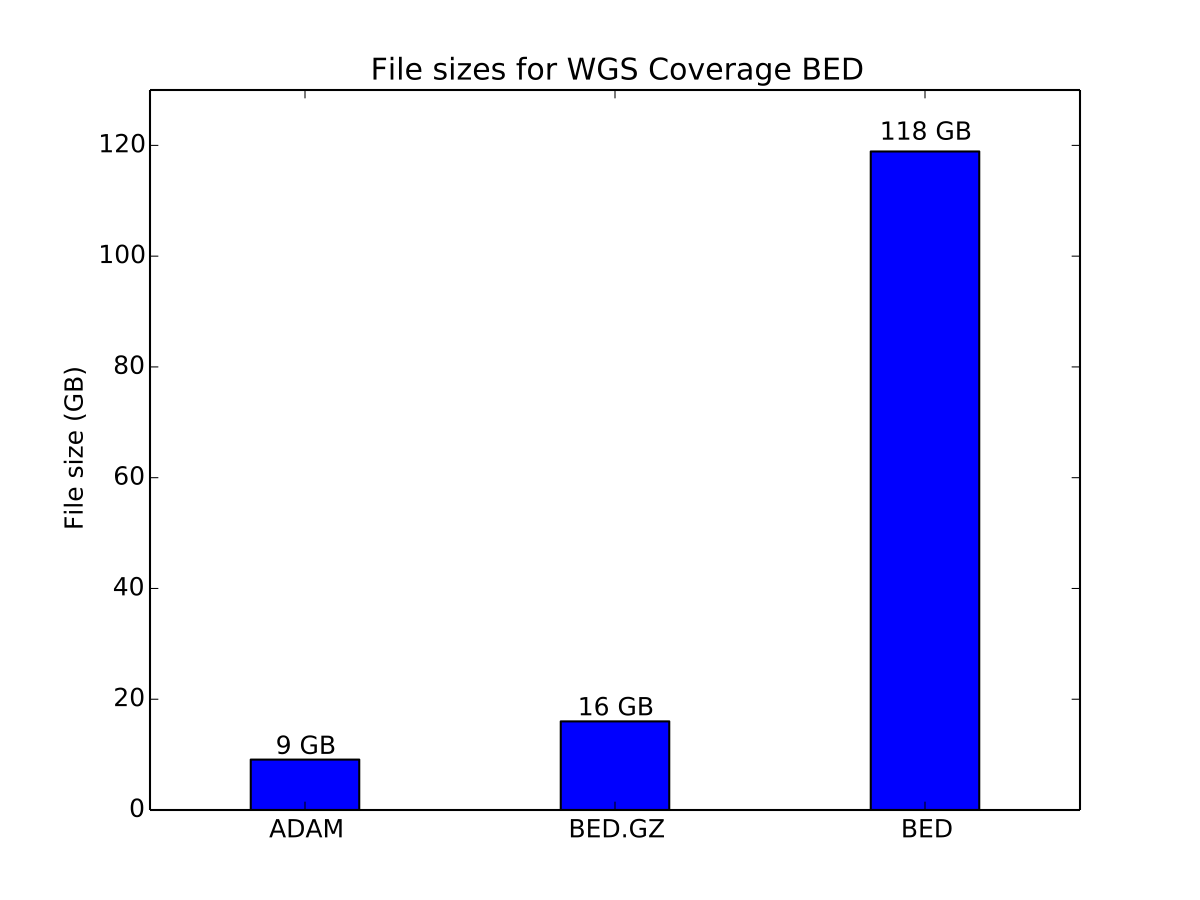
Storage cost of coverage data
For the coverage data, ADAM provides a 45% improvement in storage size relative to the compressed BED file.
Genomic Variants¶
In this benchmark, we used the 1,000 Genomes phase 3 data release VCFs. We compared GZIP-compressed VCF and uncompressed VCF to ADAM.
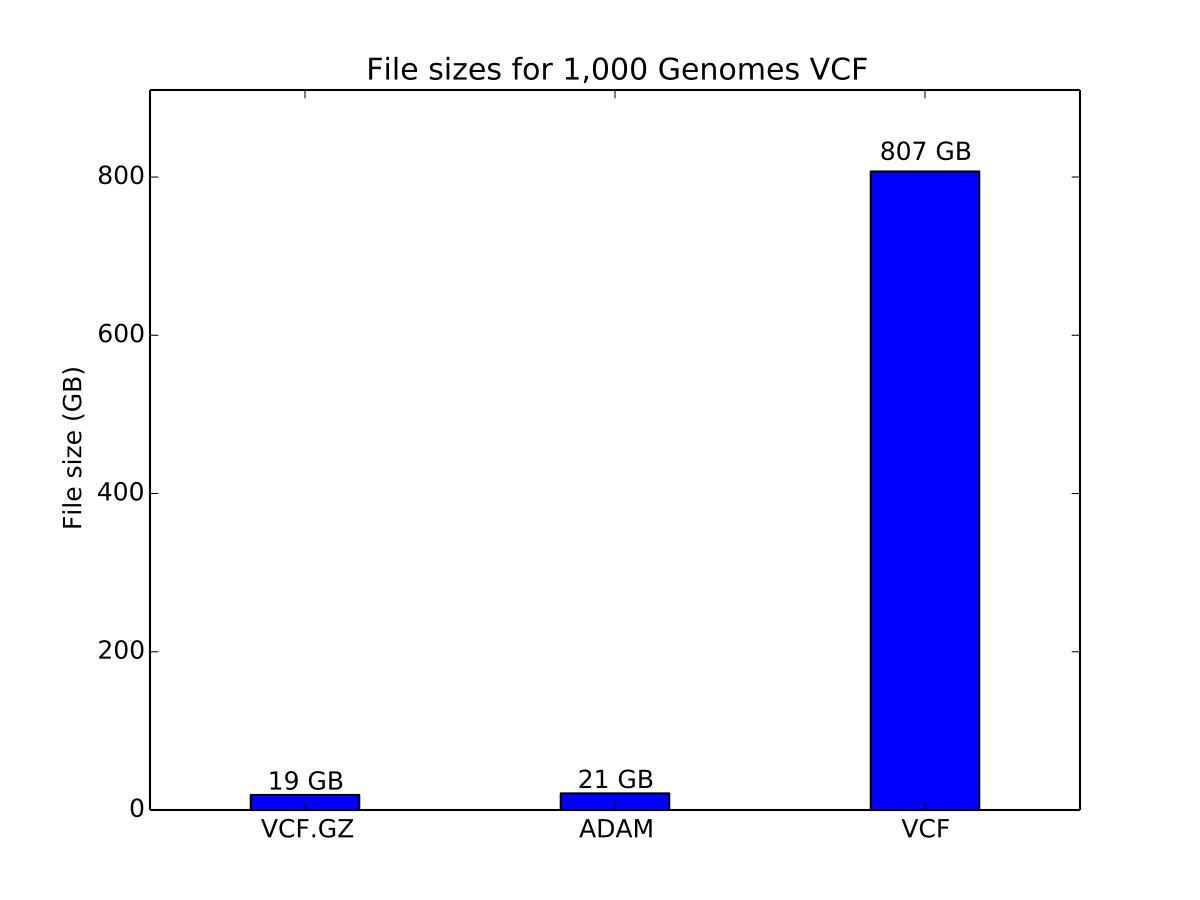
Storage cost of variant data
Compressed VCF is approximately 10% smaller than genotype data stored as Parquet.